

Judy Davie received her B.A. degree in Molecular Biology from Auburn University in 1992 and her Ph.D. degree from the University of California, Berkeley in 1998. She was awarded a postdoctoral fellowship from the American Cancer Society to study at the U.T.M.D. Anderson Cancer Center and joined SIUC in August 2006. She previously served as the Program Director for the Molecular Biology, Microbiology and Biochemistry Graduate Program.
(618) 453-5002
email: jdavie@siumed.edu
Research in my lab is focused on gene regulatory mechanisms in a development system. We focus on the specification and differentiation of skeletal muscle which is controlled by four highly related basic helix loop helix proteins referred to as the Myogenic Regulatory Factors, or MRFs. Much of the work in the lab is focused on understanding the function and specificity of myogenin, the only MRF singly required for viability. Our research is directed towards understanding the regulation of the genes controlled by myogenin by determining promoter elements that control expression and identifying additional protein factors that contribute to the unique role of myogenin on these genes, including chromatin modifying factors and additional transcription activators and co-activators.
We are also interested in how cytokine signaling pathways impact gene expression
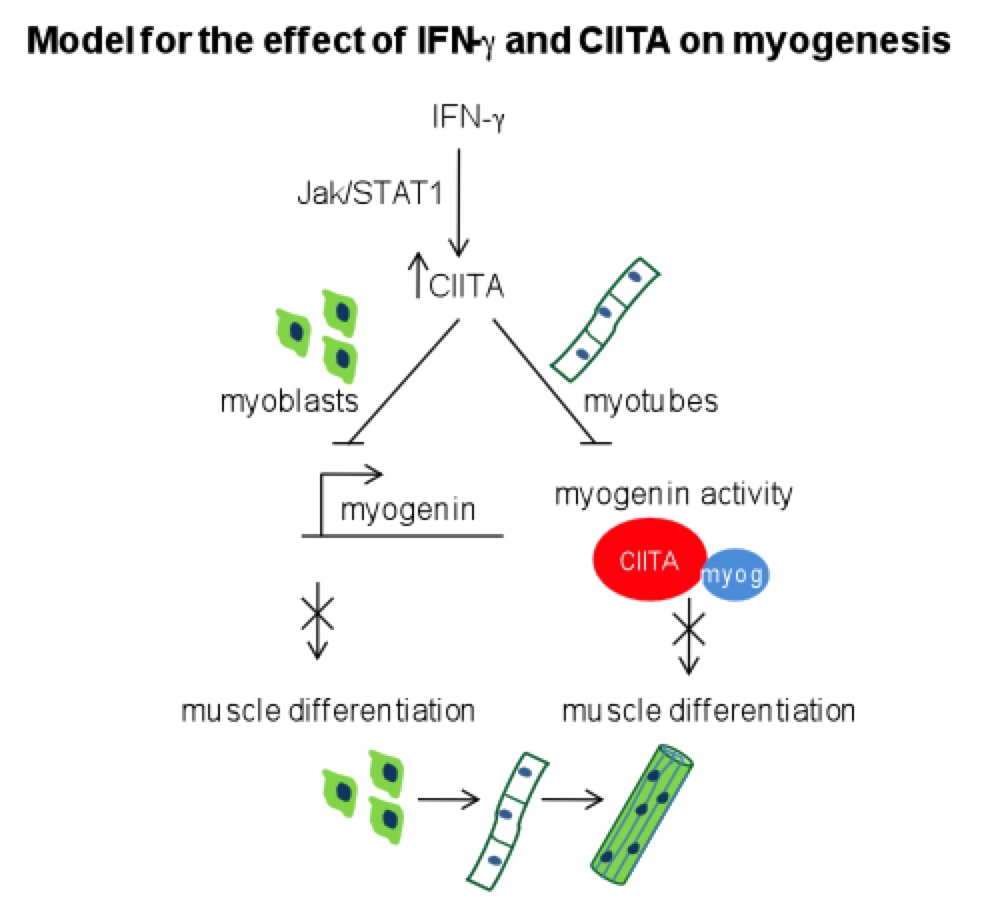
in skeletal muscle. We have shown that the inflammatory cytokine IFN-![]() ,
which was known to modulate myogenesis and be required for efficient muscle
repair, stimulates expression of CIITA in skeletal muscle cells. We have shown
that IFN-
,
which was known to modulate myogenesis and be required for efficient muscle
repair, stimulates expression of CIITA in skeletal muscle cells. We have shown
that IFN-![]() reversibly inhibits the expression and activity of myogenin through CIITA (shown
below). The lab is also investigating how IFN-
reversibly inhibits the expression and activity of myogenin through CIITA (shown
below). The lab is also investigating how IFN-![]() and CIITA contribute to efficient muscle maintenance and repair. Several muscle
diseases, including muscular dystrophy, exhibit high levels of IFN-
and CIITA contribute to efficient muscle maintenance and repair. Several muscle
diseases, including muscular dystrophy, exhibit high levels of IFN-![]() and it is highly significant to determine how CIITA contributes to disease pathology
and progression as the immune modulatory and repressive properties of IFN-
and it is highly significant to determine how CIITA contributes to disease pathology
and progression as the immune modulatory and repressive properties of IFN-![]() and CIITA may be harnessed therapeutically for skeletal muscle disorders.
and CIITA may be harnessed therapeutically for skeletal muscle disorders.
Coupled with these studies is our research to decipher how the normal process
of myogenesis is disrupted in rhabdomyosarcoma cells, a pediatric cancer. We
are interested in understanding the molecular defects in these cells to understand
how to potentially disrupt cancer growth. We have discovered that a family of
transcription factors known to promote myogenesis is deregulated in rhabdomyosarcoma
(RMS) cells which suggests that modulating the expression of these factors may
be a candidate for differentiation therapy of RMS cells. We have also found
that CIITA, the MHC class II transactivator, plays a role in the pathology of
rhabdomyosarcoma cells. We have found that RMS cells do not activate the major
histocompatibility complex genes in response to IFN-![]() .
Silencing of class II expression may allow the cancer cells to avoid detection
by the immune system and we have found that class II genes can be reactivated
by epigenetic modifiers, suggesting that this discovery could have therapeutic
potential for this pediatric cancer.
.
Silencing of class II expression may allow the cancer cells to avoid detection
by the immune system and we have found that class II genes can be reactivated
by epigenetic modifiers, suggesting that this discovery could have therapeutic
potential for this pediatric cancer.
Londhe, P., Zhu, B., Abraham, J., Keller, C. and Davie, J.K. CIITA is silenced by epigenetic mechanisms that prevent the recruitment of transactivating factors in rhabdomyosarcoma cells. International Journal of Cancer. 2011 Oct 11. epub ahead of print.
Londhe, P. and Davie, J.K. Gamma interferon inhibits myogenesis through the class II transactivator, CIITA. Molecular and Cellular Biology. 2011. Vol. 31 (14):2854-66.
Londhe, P. and Davie, J.K. Differential Recruitment of MRFs and E proteins at Differentiation Specific Genes. Skeletal Muscle Journal. 2011, Vol. 1:14.
Zhang, S., P. Londhe, M. Zhang, and Davie, J.K. Transcriptional analysis of the titin cap gene. Molecular Genetics & Genomics. 2011. Vol. 285:261-72.
Davie, J.K., Cho, J.H., Meadows, J.E., Flynn, J.M., Knapp, J.R., and Klein, W.H. Target gene selectivity of the myogenic basic helix-loop-helix transcription factor myogenin in embryonic muscle. Developmental Biology. 2007. Vol. 311 (2):650-664.
Knapp J.R., Davie J.K., Myer A., Meadows E., Olson E.N., Klein W.H. Loss of myogenin in postnatal life leads to normal skeletal muscle but reduced body size. Development. 2006. Feb;133(4):601-10. (PubMed)
Davie, J.K. and Dent, S.Y.R. No Spt6, no nucleosomes, no activator required. Molecular Cell. 2006 Feb 17;21(4):452-3.
Davie, J.K., Edmondson D.G., Coco C.B., and Dent, S.Y.R. Tup1-Ssn6 Interacts with Multiple Class I Histone Deacetylases In Vivo. Journal of Biological Chemistry. 2003. Dec 12;278(50):50158-62.
Mukai, Y., Davie, J.K., and Dent, S.Y.R. Physical and functional interaction of the yeast corepressor Tup1 with mRNA 5'-triphosphatase. Journal of Biological Chemistry. 2003 May 23;278(21):18895-901.
Edmondson, D.G., Davie, J.K., Zhou, J., Mirnikjoo, B., Tatchell, K., Dent, S.Y.R. Site-specific loss of acetylation upon phosphorylation of histone H3. Journal of Biological Chemistry. 2002 Aug 16;277(33): 29496-502.
Davie, J.K., Trumbly, R.J., Dent, S.Y.R. Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Molecular and Cell Biology. 2002 Feb;22(3):693-703.
Davie, J.K. and Dent, S.Y.R. Transcriptional control: an activating role or arginine methylation. Current Biology. 2002 Jan 22;12(2):R59-61.
Briggs, S.D., Bryk, M., Strahl, B.D., Cheung, W.L., Davie, J.K., Dent, S.Y.R, Winston, F., Allis, C.D. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes and Development. 2001 Dec 15;15(24):3286-95
Davie, J.K. and Kane, C.M. Genetic interactions between TFIIS and the Swi-Snf chromatin-remodeling complex. Molecular and Cell Biology. 2000 Aug;20(16):5960-73.
![]() Biochemistry and Molecular
Biology Home Page
Biochemistry and Molecular
Biology Home Page